

|
I PLOTName:
An I plot is used in two ways:
For both applications, the resulting plot is similar in appearance. Namely, a target value which typically appears as an X, a vertical bar with small horizontal bars at the extremes (hence the name "I plot"). The I plot has five components (characters and lines) which can be individually controlled. These components are:
For the default I plot, the I PLOT command can be preceded by the following two commands:
LINES I PLOT With these commands, the first three lines are set to blank and lines four and five are set blank. The second character is set to "X", characters one and three are set to "-", and characters four and five are set to blank. After the I plot is formed, the analyst should redefine plot characters and lines via the usual CHARACTERS and LINES commands. As mentioned above, the I plot can use location statistics other than the median. Specifically, you can use the MEAN I PLOT, MIDMEAN I PLOT, MIDRANGE I PLOT, TRIMMED MEAN I PLOT, and BIWEIGHT I PLOT to specify the mean, midmean, midrange, trimmed mean, and biweight location, respectively. This plot is also useful for displaying confidence intervals as well as other types of intervals. The following are currently supported:
where <stat> is MEDIAN, MEAN, MIDMEAN, MIDRANGE, BIWEIGHT, or TRIMMED MEAN; <y> is the response (= dependent) variable; <x> is the horizontal axis (= independent) variable; and where the <SUBSET/EXCEPT/FOR qualification> is optional. For this syntax, the extreme values will be the minimum and maximum values. If <stat> is omitted, the location statistic will be the median.
where <stat> is one of: MEAN CONFIDENCE LIMIT TRIMMED MEAN CONFIDENCE LIMIT MEDIAN CONFIDENCE LIMIT QUANTILE CONFIDENCE LIMIT BIWEIGHT CONFIDENCE LIMIT STANDARD DEVIATION CONFIDENCE LIMIT NORMAL TOLERANCE LIMIT NORMAL PREDICTION LIMIT AGRESTI COULL LIMIT ONE STANDARD ERROR TWO STANDARD ERROR ONE STANDARD DEVIATION TWO STANDARD DEVIATIONS STANDARD DEVIATION CONFIDENCE LIMIT COEFFICIENT OF VARIATION CONFIDENCE LIMIT COEFFICIENT OF DISPERSION CONFIDENCE LIMIT <y> is the response (= dependent) variable; <x> is the horizontal axis (= independent) variable; and where the <SUBSET/EXCEPT/FOR qualification> is optional. For this syntax, the extreme values will be the lower and upper bounds of the appropriate limits.
where <stat> is one of: DIFFERENCE OF MEANS CONFIDENCE LIMIT DIFFERENCE OF PROPORTIONS CONFIDENCE LIMIT RATIO OF MEANS CONFIDENCE LIMIT CORRELATION CONFIDENCE LIMIT <y1> is the first response (= dependent) variable; <y2> is the second response (= dependent) variable; <x> is the horizontal axis (= independent) variable; and where the <SUBSET/EXCEPT/FOR qualification> is optional. This is similar to Syntax 2 except that there are two response variables instead of one.
<SUBSET/EXCEPT/FOR qualification> where <stat> is one of: MEDIAN I MEAN I MIDMEAN I MIDRANGE I BIWEIGHT I TRIMMED MEAN I MEAN CONFIDENCE LIMIT TRIMMED MEAN CONFIDENCE LIMIT MEDIAN CONFIDENCE LIMIT QUANTILE CONFIDENCE LIMIT BIWEIGHT CONFIDENCE LIMIT STANDARD DEVIATION CONFIDENCE LIMIT COEFFICIENT OF VARIATION CONFIDENCE LIMIT COEFFICIENT OF DISPERSION CONFIDENCE LIMIT NORMAL TOLERANCE LIMIT NORMAL PREDICTION LIMIT AGRESTI COULL LIMIT ONE STANDARD ERROR TWO STANDARD ERROR ONE STANDARD DEVIATION TWO STANDARD DEVIATION <y1> ... <yk> is a list of up to k response variables; and where the <SUBSET/EXCEPT/FOR qualification> is optional. This syntax is useful when the data for each group is stored in separate response variables. Up to 30 response variables may be specified. Note that the syntax
is supported. This is equivalent to
<SUBSET/EXCEPT/FOR qualification> where <stat> is one of the variants listed for Syntax 4; <y> is the response variable; <x1> ... <xk> is a list of up to k group-id variables; and where the <SUBSET/EXCEPT/FOR qualification> is optional. This syntax performs a cross-tabulation of <x1> ... <xk> and generates an i plot (on the same graph) for each unique combination of cross-tabulated values. For example, if X1 has 3 levels and X2 has 2 levels, there will be a total of 6 i plots generated. Up to six group-id variables can be specified. Note that the syntax REPLICATED I PLOT Y X1 TO X4 is supported. This is equivalent to REPLICATED I PLOT Y X1 X2 X3 X4 In this syntax, all i plots are drawn with the sampe plot attributes. As an example of how the x-coordinates are determined, assume we have 3 levels for X1 and 2 levels for X2. The x-coordinates will be
<SUBSET/EXCEPT/FOR qualification> where <stat> is one of the variants listed for Syntax 4; <y> is the response variable; <x1> is the first group-id variable; <x2> is the second group-id variable; and where the <SUBSET/EXCEPT/FOR qualification> is optional. This syntax is similar to syntax 5. However, it has the following distinctions:
Program 2 demonstrates the difference between Syntax 5 and Syntax 6.
I PLOT Y X SUBSET X > 2 ONE STANDARD ERROR PLOT Y X MEAN CONFIDENCE LIMIT PLOT Y X
To specify the alpha level for the confidence limit cases, enter the command (0.05 will be used if this command is not given)
For the QUANTILE CONFIDENCE LIMIT case, specify the desired quantile with the command (0.50 will be used if this command is not given)
For the TRIMMED MEAN CONFIDENCE LIMIT case, specify the trimming parameters with the commands (0.25 and 0.75 will be used if these commands are not given)
LET P2 = <value>
<WILSON/ADJUSTED WALD/JEFFREYS/NORMAL/EXACT> The default is the Wilson method. For details on the various methods, enter HELP PORPORTION CONFIDENCE LIMITS. The difference of proportion confidence limits also support several different methods for deteriming the confidence limits. To specify the method to use, enter the command To specify the method to use, enter the command
<WALD/ADJUSTED WALD/BAYESIAN> The default is the adjusted Wald (Agresti-Caffo) interval. For details on the various methods, enter HELP DIFFERENCE OF PORPORTION CONFIDENCE LIMITS.
2011/02: Support for MULTIPLE and REPLICATION options 2013/10: Support for MEAN, MIDMEAN, MIDRANGE I plots 2013/10: Support for various confidence interval cases (Syntax 2) 2013/10: Support for special 3 variable case (Syntax 5) 2017/11: Support for DIFFERENCE OF MEANS CONFIDENCE LIMIT 2017/11: Support for DIFFERENCE OF PROPORTIONS CONFIDENCE LIMIT 2017/11: Support for COEFFICIENT OF VARIATION CONFIDENCE LIMIT 2017/11: Support for COEFFICIENT OF DISPERSION CONFIDENCE LIMIT 2017/12: Support for CORRELATION CONFIDENCE LIMIT 2019/10: Support for RATIO OF MEANS CONFIDENCE LIMIT
. Step 1: Read the data
.
skip 25
read gear.dat y x
.
. Step 2: Basic i-plot
.
title case asis
title offset 2
label case asis
character case asis
line i plot
character blank circle blank blank blank
character fill off on
character hw 0.5 0.375 all
xlimits 1 10
major xtic mark number 10
minor xtic mark number 0
x1tic mark offset 0.5 0.5
title automatic
.
multiplot corner coordinates 5 5 95 95
multiplot scale factor 2
multiplot 2 2
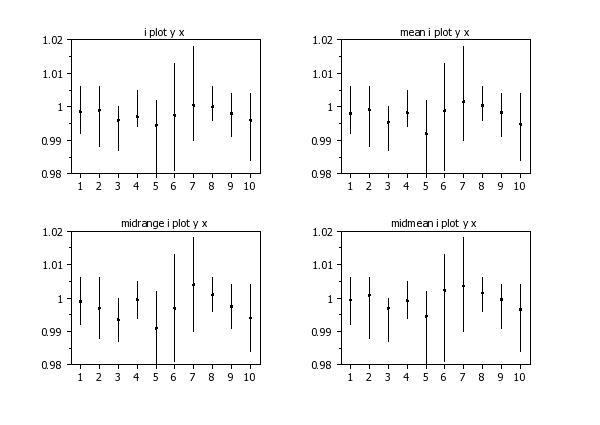
i plot y x
mean i plot y x
midrange i plot y x
midmean i plot y x
end of multiplot
.
multiplot 2 2
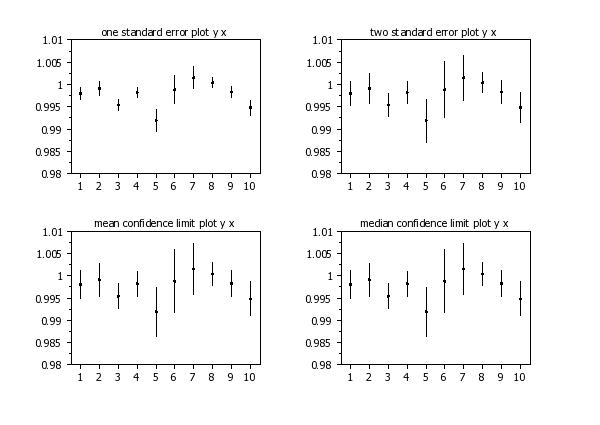
one standard error plot y x
two standard error plot y x
mean confidence limit plot y x
median confidence limit plot y x
end of multiplot
.
multiplot 2 2
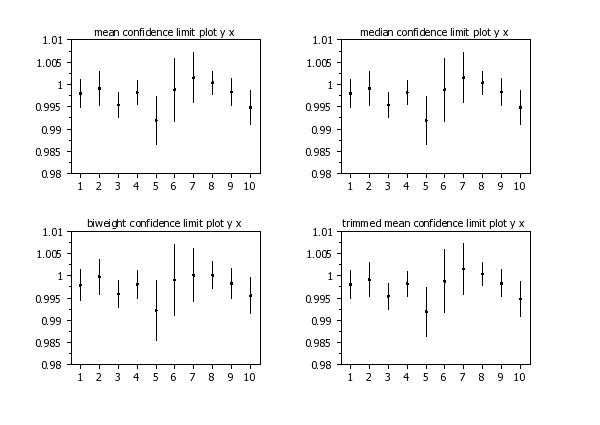
mean confidence limit plot y x
median confidence limit plot y x
biweight confidence limit plot y x
trimmed mean confidence limit plot y x
end of multiplot
 Program 2:
Program 2:
. Step 1: Read the data
.
. y = voltage drop
. x1 = elapsed days (1, 8, 15, 29, 43, 57, 71, 107)
. x2 = type of connector
. 1 = brass
. 2 = steel
. 3 = innovative
skip 25
read clark0.dat y days x2
let days = 8 subset days = 9
let days = 15 subset days = 16
let days = 29 subset days = 30
let days = 43 subset days = 44
let days = 57 subset days = 58
let days = 71 subset days = 72
let days = 107 subset days = 108
let x1 = coded days
let nx1 = unique x1
let nx2 = unique x2
let ntot = nx1*nx2
.
. Step 2: I Plot with standard replication option
.
title case asis
title offset 2
label case asis
character case asis
line i plot
character blank circle blank blank blank
character fill off on
character hw 0.5 0.375 all
.
y1label Voltage Drop
x3label Elapsed Days/Connector Type
xlimits 1 ntot
major xtic mark number ntot
minor xtic mark number 0
x1tic mark offset 0.5 0.5
x1tic mark label format alpha
x1tic mark label content 1 8 15 29 43 57 71 107 1 8 15 29 43 57 71 107 ...
1 8 15 29 43 57 71 107
x1tic mark label size 1.6
title automatic
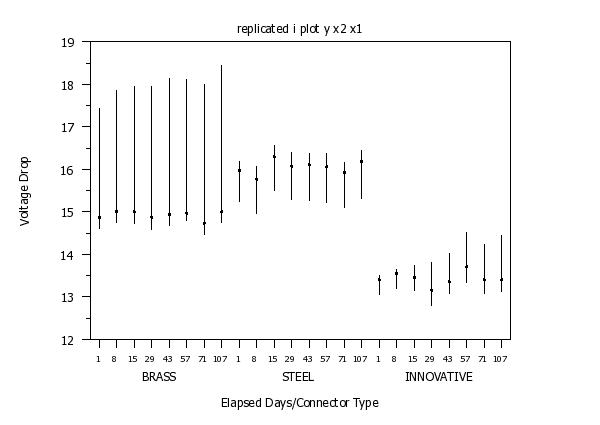
replicated i plot y x2 x1
.
justification center
let xcoor = (nx1/2) + 0.5
let ycoor = 10
moveds xcoor ycoor
text Brass
let xcoor = xcoor + nx1
moveds xcoor ycoor
text Steel
let xcoor = xcoor + nx1
moveds xcoor ycoor
text Innovative
.
. Step 3: I Plot with alternative replication option
.
line i plot bl bl bl so so bl bl bl so so bl bl bl so so
character bl circ bl bl bl bl circ bl bl bl bl circ bl bl bl
character fill on all
character hw 0.5 0.375 all
character color blue blue blue blue blue red red red red red ...
green green green green green
line color blue blue blue blue blue red red red red red ...
green green green green green
.
x1label Elapsed Days/Connector Type
x3label
xlimits 1 nx1
major xtic mark number nx1
minor xtic mark number 0
x1tic mark offset 0.5 0.5
x1tic mark label format alpha
x1tic mark label content 1 8 15 29 43 57 71 107
title automatic
i plot y x1 x2
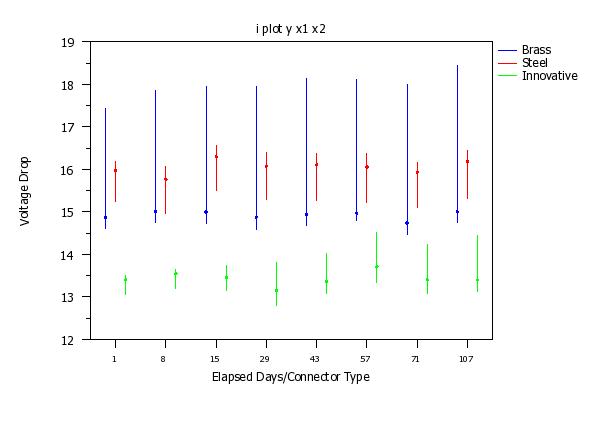 Program 3:
Program 3:
. Step 1: Read the data
.
. x1 = instrument
. x2 = source
. y1 = expected alarm
. y2 = observed alarm
. y = 1 if y1 and y2 agree, 0 otherwise
skip 25
read alarm.dat x1 x2 y1 y2
skip 0
let n = size y1
let y = 0 for i = 1 1 n
let y = 1 subset y1 = 0 subset y2 = 0
let y = 1 subset y1 = 1 subset y2 = 1
let ninst = unique x1
let nsrc = unique x2
.
.
. Step 2: I Plot with standard replication option
.
line i plot
character blank circle blank blank blank
character fill off on
character hw 0.5 0.375 all
.
label case asis
title case asis
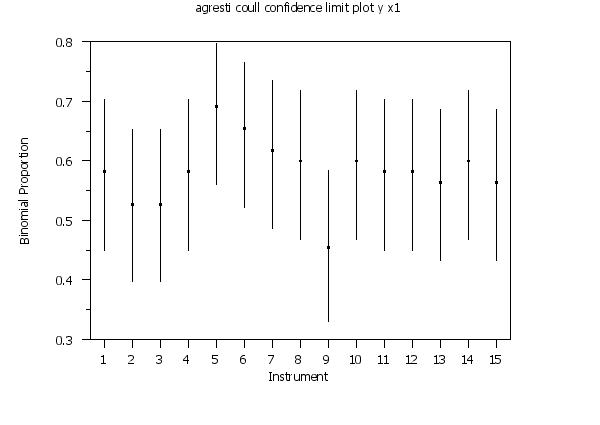
y1label Binomial Proportion
x1label Instrument
xlimits 1 ninst
major xtic mark number ninst
minor xtic mark number 0
x1tic mark offset 0.5 0.5
title automatic
agresti coull confidence limit plot y x1
.
difference of proportion confidence limit plot y1 y2 x1
Date created: 11/05/2013 |
Last updated: 12/04/2023 Please email comments on this WWW page to [email protected]. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||