

|
HOMOSCEDASTICITY PLOTName:
The interpretation of this plot is that the greater the spread on the vertical axis, the less valid is the assumption of constant variance. By default, this command assumes that you have raw data. In some cases, you may only have summary data (i.e., means and standard deviations or some other location/scale statistics). See Syntax 4 and Syntax 5 for how to use this command with summary data. You can also use this command when you have more than one group-id variable.
<SUBSET/EXCEPT/FOR qualification> where <y> is a response variable; <x1> ... <xk> is a list of one to six group-id variables; and where the <SUBSET/EXCEPT/FOR qualification> is optional. From one to six group-id variables can be specified (most commonly there is a single group-id variable). Note that with this syntax, all plot points are drawn with the same characteristics (i.e., the first setting of the CHARACTER and LINE commands and their associated attribute setting commands).
<SUBSET/EXCEPT/FOR qualification> where <y> is a response variable; <x1> ... <xk> is a list of one to six group-id variables; and where the <SUBSET/EXCEPT/FOR qualification> is optional. From one to six group-id variables can be specified (most commonly there is a single group-id variable). Note that with this syntax, the plot points corresponding to each subset are drawn with different attributes (i.e., the first subset uses the first setting for the CHARACTER and LINE and related attribute setting commands, the second subset uses the second setting, and so on). For example, this syntax can be used to label the plot points with the lab-id. If there is more than one group-id variable, the attribute settings work from right to left. That is, if X1 has 2 levels and X2 has 2 levels, then
<SUBSET/EXCEPT/FOR qualification> where <y1> ... <yk> is a list of one to 30 response variables; and where the <SUBSET/EXCEPT/FOR qualification> is optional. With this syntax, the different subsets are represented with different response variables. As with Syntax 2, each subset is drawn with different attributes.
<SUBSET/EXCEPT/FOR qualification> where <ymean> is a variable containing subset means; <ysd> is a variable containing subset standard deviations; <nrepl> is an optional variable containing the number of replications in each subset; and where the <SUBSET/EXCEPT/FOR qualification> is optional. This syntax is used when you only have summary data. The <ymean> variable contains the means (or some other location statistic) and <ysd> contains the standard deviations (or some other scale statistic) for the different subsets. The <nrepl> variable is only used when the CIRCLE TECHNIQUE is used to generate lines of "similar homogeneity". See the Note section below for a description of the CIRCLE TECHNQUE. As with Syntax 1, all plot points are drawn with the same attributes.
<SUBSET/EXCEPT/FOR qualification> where <ymean> is a variable containing subset means; <ysd> is a variable containing subset standard deviations; <nrepl> is an optional variable containing the number of replications in each subset; and where the <SUBSET/EXCEPT/FOR qualification> is optional. This syntax is used when you only have summary data. The <ymean> variable contains the means (or some other location statistic) and <ysd> contains the standard deviations (or some other scale statistic) for the different subsets. The <nrepl> variable is only used when the CIRCLE TECHNIQUE is used to generate lines of "similar homogeneity". See the Note section below for a description of the CIRCLE TECHNQUE. As with Syntax 2, the different subsets are plotted with different attributes.
HOMOSCEDASTICITY PLOT Y1 TAG SUBSET TAG > 2 MULTIPLE HOMOSCEDASTICITY PLOT Y1 Y2 Y3 Y4 Y5 MULTIPLE HOMOSCEDASTICITY PLOT Y1 TO Y5 SUBSET HOMOSCEDASTICITY PLOT Y1 TAG SUMMARY SUBSET HOMOSCEDASTICITY PLOT YMEAN YSD SUBSET HOMOSCEDASTICITY PLOT Y1 X1 X2
You can set the location statistic with the commands
SET HOMOSCEDASTICITY LOCATION BIWEIGHT LOCATION SET HOMOSCEDASTICITY LOCATION H10 SET HOMOSCEDASTICITY LOCATION H12 SET HOMOSCEDASTICITY LOCATION H15 SET HOMOSCEDASTICITY LOCATION H17 SET HOMOSCEDASTICITY LOCATION H20 SET HOMOSCEDASTICITY LOCATION HODGES LEHMAN SET HOMOSCEDASTICITY LOCATION LP LOCATION SET HOMOSCEDASTICITY LOCATION MEDIAN SET HOMOSCEDASTICITY LOCATION MIDMEAN SET HOMOSCEDASTICITY LOCATION MIDRANGE SET HOMOSCEDASTICITY LOCATION TRIMMED MEAN SET HOMOSCEDASTICITY LOCATION WINSORIZED MEAN You can set the scale statistic with the commands
SET HOMOSCEDASTICITY SCALE BIWEIGHT SCALE SET HOMOSCEDASTICITY SCALE H10 SET HOMOSCEDASTICITY SCALE H12 SET HOMOSCEDASTICITY SCALE H15 SET HOMOSCEDASTICITY SCALE H17 SET HOMOSCEDASTICITY SCALE H20 SET HOMOSCEDASTICITY SCALE AVERAGE ABSOLUTE DEVIATION (or AAD) SET HOMOSCEDASTICITY SCALE MEDIAN ABSOLUTE DEVIATION (or MAD) SET HOMOSCEDASTICITY SCALE INTERQUARTILE RANGE (or IQ) SET HOMOSCEDASTICITY SCALE RANGE SET HOMOSCEDASTICITY SCALE TRIMMED SD SET HOMOSCEDASTICITY SCALE WINSORIZED SD SET HOMOSCEDASTICITY SCALE SN SET HOMOSCEDASTICITY SCALE QN
The method used is referred to as the "circle technique". To generate these confidence contours, enter the command
Confidence contours are drawn at \( \alpha \) = 0.95, 0.99, and 0.999. The first three settings of the LINE and CHARACTER commands (and related attribute setting commands) are used to generate these confidence contours. The LINE and CHARACTER settings for the plot points start with trace 4. Dataplot uses the method as given in ISO 13528. This is a slight variation of the method originally given by van Nuland. This method is typically applied in the context of proficiency testing. It assumes that there are p labs with each lab having n replicate measurements. The mean and standard deviation are computed for each lab. We then compute a pooled mean, \( \bar{X} \), and a pooled standard deviation, \( \bar{S} \), for the p labs. The pooled mean is computed from the p means using the H15 location estimate. This is referred to as Algorithm A in the 13528 standard. The pooled standard deviation is computed using Algorithm S as defined in the 13528 standard. The (x,y) values for a critical region with significance level \( \alpha \) can be computed from
and
with \( \chi^{2} \) and n denoting the chi-square percent point function and the number of replicated measurements per lab, respectively. If the number of replications is not equal for the labs, Dataplot will use an average number of replications. Dataplot follows the ISO-13528 standard in computing these curves for \( \alpha \) = 95%, 99%, and 99.9%.
for details.
HIGHLIGHT HOMOSCEDASTICITY is a synonym for SUBSET HOMOSCEDASTICITY
The terms SUMMARY, SUBSET, MULTIPLE and HOMOSCEDASTICITY can
be entered in arbitrary order on the command.
van Nuland (1992), "ISO 9002 and the circle technique," Qual. Eng., 5, pp. 269-291.
2010/12: Support for MULTIPLE and SUBSET/HIGHLIGHT option 2010/12: Support for SUMMARY option 2010/12: Support for alternate location and scale statistics 2010/12: Support for "circle technique" for identifying labs that are statistically significantly different 2010/12: Support for more than one group-id variable
skip 25
read gear.dat y x
.
title case asis
title offset 2
label case asis
x1label Mean
y1label Standard Deviation
xlimits 0.990 1.005
ylimits 0 0.01
ytic mark offset 0 0.002
major ytic mark number 6
minor ytic mark number 1
y1label displacement 12
.
line blank all
char x all
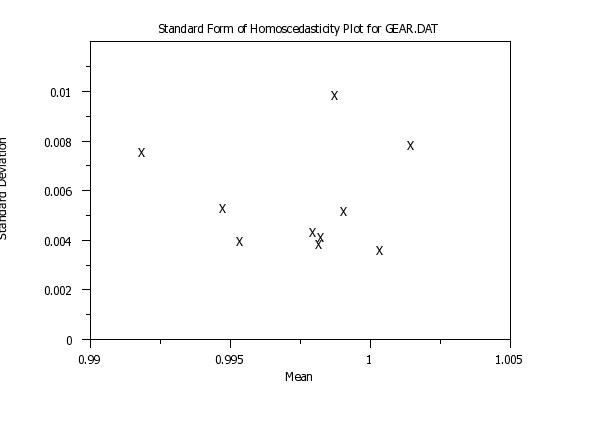
title Standard Form of Homoscedasticity Plot for GEAR.DAT
homoscedasticity plot y x
.
char 1 2 3 4 5 6 7 8 9 10
title Highlight Form of Homoscedasticity Plot for GEAR.DAT
highlight homoscedasticity plot y x
.
. Different Statistics
.
title Different Location/Scale Statistics for GEAR.DAT
label
yminimum 0
subset homoscedasticity plot y x
pre-erase off
set homo plot location h15
set homo plot scale h15
x1label H15 Location
y1label H15 Scale
char color red all
subset homoscedasticity plot y x
pre-erase on
skip 25
read ripken.dat y x1 to x4
.
title case asis
label case asis
x1label Mean
y1label Standard Deviation
line blank all
.
char 1 2 3 1 2 3 1 2 3
char color red red red blue blue blue green green green
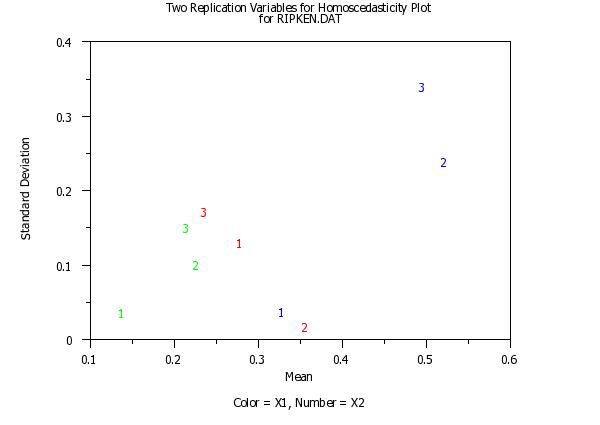
x3label Color = X1, Number = X2
title Two Replication Variables for Homoscedasticity Plotcr() ...
for RIPKEN.DAT
homoscedasticity plot y x1 to x2
 Program 3:
Program 3:
SKIP 25
READ MONTGOME.DAT Y1 Y2 Y3
.
title case asis
label case asis
x1label Mean
y1label Standard Deviation
line blank all
.
char 1 2 3
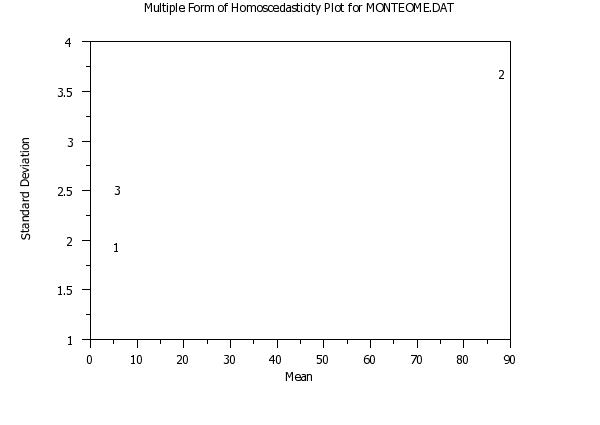
title Multiple Form of Homoscedasticity Plot for MONTEOME.DAT
multiple homoscedasticity plot y1 y2 y3
 Program 4:
Program 4:
skip 25
read gear.dat y x
.
title case asis
label case asis
x1label Mean
y1label Standard Deviation
line blank all
.
char blank blank blank 1 2 3 4 5 6 7 8 9 10
line solid dash dotted
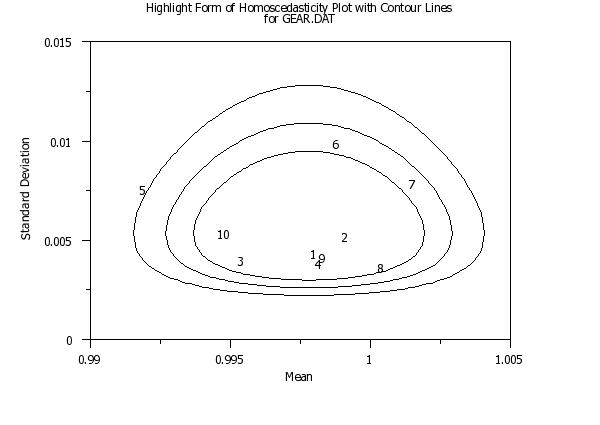
title Highlight Form of Homoscedasticity Plot with Contour Linescr() ...
for GEAR.DAT
.
xlimits 0.99 1.005
ylimits 0 0.015
set homo plot circle technique on
pre-sort off
highlight homoscedasticity plot y x
pre-sort on
 Program 5:
Program 5:
skip 25
read antibody.dat lab ymean ysd nrepl
.
title case asis
label case asis
x1label Mean
y1label Standard Deviation
.
char 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 ...
19 20 21 22 23 24 25
line blank all
.
title Highlight/Summary Form of Homoscedasticity Plot with Contour Linescr() ...
for ANTIBODY.DAT
highlight summary homoscedasticity plot ymean ysd
.
char blank blank blank 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 ...
19 20 21 22 23 24 25
line blank all
line solid dash dotted
xlimits 0.8 2.5
ylimits 0 1.6
.
title Highlight/Summary Form of Homoscedasticity Plot with Contour Linescr() ...
for ANTIBODY.DAT
set homo plot circle technique on
highlight summary homoscedasticity plot ymean ysd nrepl
Date created: 12/06/2010 |
Last updated: 12/04/2023 Please email comments on this WWW page to [email protected]. | ||||||||||||||||||||||||||||||||||||||||||