

|
E691 INTERLABName:
describes a methodology for performing an interlab experiment. The data consists of:
It is assumed that this is a balanced design. That is, each combination of lab-id and material-id (referred to as a cell in the standard) has an equal number of observations. The purpose of the E691 INTERLAB command is to estimate the precision of a test method. Two important concepts in determining the precision are:
The standard discusses issues regarding the design of the interlaboratory study. We do not dicuss those issues here. If your data is stored in a format with each lab in a separate column, you can use the REPLICATED STACK command to save your data in the format needed by the E691 INTERLAB command. This command generates the four tables documented in the above document:
The following provides some of the quantities that are computed in this command (see the above document for a more detailed discussion of these quantities).
<SUBSET/EXCEPT/FOR qualification> where <y> is a response variable; <labid> is a lab id variable; <matid> is a material id variable; and where the <SUBSET/EXCEPT/FOR qualification> is optional.
E691 INTERLAB Y1 LABID MATID SUBSET LABID > 1
The following graphs are recommended in the ASTM document:
We also recommend the following graphs:
We have added the following macros to Dataplot's built-in macro library:
To call one of these macros, define the following values
LABID = lab-id variable MATID = material-id variable E691TIT = string containing a title for the analysis and then use the CALL command to invoke the macro. This is demonstrated in the program example below. If you would like to view one of these macros, use the LIST command. For example
If you would like to modify one of these macros, use the COPY command to create a copy of the macro. For example
You can then edit C:\E691PLO1_NEW.DP to suit your needs.
The second program example demostrates this for Latex.
SET E692 TEST RESULTS TABLE <ON/OFF> SET E692 H CONSISTENCY TABLE <ON/OFF> SET E692 K CONSISTENCY TABLE <ON/OFF> SET E692 PRECISION STATISTICS TABLE <ON/OFF>
If you have defined a group label variable (HELP GROUP LABEL for details) that defines alphabetic codes for the material names, you can use the command
where <group-id variable> is the name of a group-id variable. To restore the use of numeric codes, enter
The 2024/05 version updated the E691 command to support unbalanced data. The formulas for the unbalanced data are given in section A2 of the 2023 version of the standard and are not given here.
2008/10: Made a number of corrections to the documentation 2015/03: SET commands to allow user control over which tables will be printed 2015/03: Added table to print raw data 2024/05: Added support for unbalanced data
DIMENSION 100 VARIABLES
SKIP 25
READ GLUCOSE.DAT Y LABID MATID
LET STRING E691TIT = Glucose in Serum
SET TABLE TITLE ^E691TIT
.
E691 INTERLAB Y LABID MATID
.
CALL E691PLO1.DP
CALL E691PLO2.DP
CALL E691PLO3.DP
CALL E691PLO4.DP
CALL E691PLO5.DP
CALL E691PLO6.DP
CALL E691PLO7.DP
The following tables are generated
INTERLABORATORY ANALYSIS (BASED ON E 691 - 99 STANDARD)
Test Result Data
-------------------------------------------------------------------------------------
Material
Laboratory 1 2 3 4 5
-------------------------------------------------------------------------------------
1 41.0300 78.2800 132.6600 193.7100 292.7800
1 41.4500 78.1800 133.8300 193.5900 294.0900
1 41.3700 78.4900 133.1000 193.6500 292.8900
2 41.1700 77.7800 132.9200 190.8800 292.2700
2 42.0000 80.3800 136.9000 200.1400 309.4000
2 41.1500 79.5400 136.4000 194.3000 295.0800
3 41.0100 79.1800 132.6100 192.7100 295.5300
3 40.6800 79.7200 135.8000 193.2800 290.1400
3 42.6600 80.8100 135.3600 190.2800 292.3400
4 39.3700 84.0800 138.5000 195.8500 295.1900
4 42.3700 78.6000 148.3000 196.3600 295.4400
4 42.6300 81.9200 135.6900 199.4300 296.8300
5 41.8800 78.1600 131.9000 192.5900 293.9300
5 41.1900 79.5800 134.1400 191.4400 292.4800
5 41.3200 78.3300 133.7600 195.1200 294.2800
6 43.2800 78.6600 137.2100 195.3400 297.7400
6 40.5000 79.2700 135.1400 198.2600 296.8000
6 42.2800 81.7500 137.5000 198.1300 290.3300
7 41.0800 79.7600 130.9700 194.6600 287.2900
7 41.2700 81.4500 131.5900 191.9900 293.7600
7 39.0200 77.3500 134.9200 187.1300 289.3600
8 43.3600 80.4400 135.4600 197.5600 298.4600
8 42.6500 80.8000 135.1400 195.9900 295.2800
8 41.7200 79.8000 133.6300 200.8200 296.1200
Glucose
Initial Preparation of Test Result Data for Material: 1
================================================================================
Laboratory Cell Cell
Number Mean SD d h k
================================================================================
1 41.2833 0.2230 -0.2350 -0.39 0.21
2 41.4400 0.4851 -0.0783 -0.13 0.46
3 41.4500 1.0608 -0.0683 -0.11 1.00
4 41.4567 1.8118 -0.0617 -0.10 1.70
5 41.4633 0.3667 -0.0550 -0.09 0.34
6 42.0200 1.4081 0.5017 0.83 1.32
7 40.4567 1.2478 -1.0617 -1.75 1.17
8 42.5767 0.8225 1.0583 1.75 0.77
================================================================================
Average of cell averages = 41.51833
Standard deviation of cell averages = 0.60613
Repeatability standard deviation = 1.06322
Reproducibility standard deviation = 1.06322
Glucose
Initial Preparation of Test Result Data for Material: 2
================================================================================
Laboratory Cell Cell
Number Mean SD d h k
================================================================================
1 78.3167 0.1582 -1.3629 -1.36 0.11
2 79.2333 1.3269 -0.4463 -0.45 0.89
3 79.9033 0.8303 0.2238 0.22 0.56
4 81.5333 2.7604 1.8538 1.85 1.85
5 78.6900 0.7754 -0.9896 -0.99 0.52
6 79.8933 1.6366 0.2138 0.21 1.09
7 79.5200 2.0605 -0.1596 -0.16 1.38
8 80.3467 0.5065 0.6671 0.67 0.34
================================================================================
Average of cell averages = 79.67958
Standard deviation of cell averages = 1.00275
Repeatability standard deviation = 1.49485
Reproducibility standard deviation = 1.57963
Glucose
Initial Preparation of Test Result Data for Material: 3
================================================================================
Laboratory Cell Cell
Number Mean SD d h k
================================================================================
1 133.1967 0.5910 -1.9462 -0.73 0.22
2 135.4067 2.1680 0.2637 0.10 0.79
3 134.5900 1.7288 -0.5529 -0.21 0.63
4 140.8300 6.6200 5.6871 2.14 2.41
5 133.2667 1.1987 -1.8763 -0.71 0.44
6 136.6167 1.2870 1.4738 0.55 0.47
7 132.4933 2.1243 -2.6496 -1.00 0.77
8 134.7433 0.9774 -0.3996 -0.15 0.36
================================================================================
Average of cell averages = 135.14291
Standard deviation of cell averages = 2.65595
Repeatability standard deviation = 2.74827
Reproducibility standard deviation = 3.47698
Glucose
Initial Preparation of Test Result Data for Material: 4
================================================================================
Laboratory Cell Cell
Number Mean SD d h k
================================================================================
1 193.6500 0.0600 -1.0671 -0.41 0.02
2 195.1067 4.6824 0.3896 0.15 1.78
3 192.0900 1.5932 -2.6271 -1.01 0.61
4 197.2133 1.9365 2.4962 0.96 0.74
5 193.0500 1.8826 -1.6671 -0.64 0.72
6 197.2433 1.6496 2.5262 0.97 0.63
7 191.2600 3.8177 -3.4571 -1.33 1.45
8 198.1233 2.4638 3.4063 1.31 0.94
================================================================================
Average of cell averages = 194.71709
Standard deviation of cell averages = 2.59500
Repeatability standard deviation = 2.62506
Reproducibility standard deviation = 3.36571
Glucose
Initial Preparation of Test Result Data for Material: 5
================================================================================
Laboratory Cell Cell
Number Mean SD d h k
================================================================================
1 293.2533 0.7267 -1.2387 -0.46 0.18
2 298.9167 9.1869 4.4246 1.64 2.33
3 292.6700 2.7101 -1.8221 -0.68 0.69
4 295.8200 0.8836 1.3279 0.49 0.22
5 293.5633 0.9544 -0.9287 -0.34 0.24
6 294.9567 4.0343 0.4646 0.17 1.03
7 290.1367 3.3042 -4.3554 -1.62 0.84
8 296.6200 1.6479 2.1279 0.79 0.42
================================================================================
Average of cell averages = 294.49207
Standard deviation of cell averages = 2.69314
Repeatability standard deviation = 3.93497
Reproducibility standard deviation = 4.19233
Glucose-h
=============================================================
Material
Laboratory 1 2 3 4 5
=============================================================
1 -0.39 -1.36 -0.73 -0.41 -0.46
2 -0.13 -0.45 0.10 0.15 1.64
3 -0.11 0.22 -0.21 -1.01 -0.68
4 -0.10 1.85 2.14 0.96 0.49
5 -0.09 -0.99 -0.71 -0.64 -0.34
6 0.83 0.21 0.55 0.97 0.17
7 -1.75 -0.16 -1.00 -1.33 -1.62
8 1.75 0.67 -0.15 1.31 0.79
=============================================================
CRITICAL VALUE = 2.15
Glucose-k
Material
Laboratory 1 2 3 4 5
=============================================================
1 0.21 0.11 0.22 0.02 0.18
2 0.46 0.89 0.79 1.78 2.33
3 1.00 0.56 0.63 0.61 0.69
4 1.70 1.85 2.41 0.74 0.22
5 0.34 0.52 0.44 0.72 0.24
6 1.32 1.09 0.47 0.63 1.03
7 1.17 1.38 0.77 1.45 0.84
8 0.77 0.34 0.36 0.94 0.42
=============================================================
CRITICAL VALUE = 2.06
Glucose-Precision Statistics
========================================================================================
Material Xbar s(x) s(r) s(R) r R
========================================================================================
1 41.5183 0.6061 1.0632 1.0632 2.98 2.98
2 79.6796 1.0028 1.4949 1.5796 4.19 4.42
3 135.1429 2.6559 2.7483 3.4770 7.70 9.74
4 194.7171 2.5950 2.6251 3.3657 7.35 9.42
5 294.4921 2.6931 3.9350 4.1923 11.02 11.74
========================================================================================
The following graphs are generated.
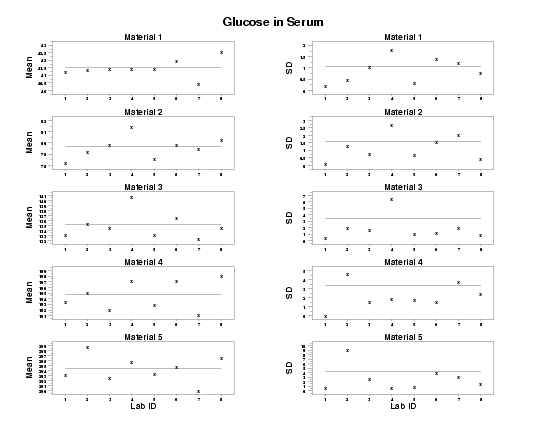
DIMENSION 100 VARIABLES
SKIP 25
READ MANDEL6.DAT Y LABID MATID
LET STRING E691TIT = Pentosans in Wood Pulp
SET TABLE TITLE ^E691TIT
.
. DEMONSTRATE HOW TO SAVE OUTPUT IN A LATEX FILE
. WITH GRAPHICS
CAPTURE LATEX E691.TEX
E691 INTERLAB Y LABID MATID
.
SET IPL1NA E691PLO1.PS
DEVICE 2 POSTSCRIPT
CALL E691PLO1.DP
DEVICE 2 CLOSE
SET IPL1NA E691PLO2.PS
DEVICE 2 POSTSCRIPT
CALL E691PLO2.DP
DEVICE 2 CLOSE
SET IPL1NA E691DOH1.PS
DEVICE 2 POSTSCRIPT
CALL E691DOH1.DP
DEVICE 2 CLOSE
SET IPL1NA E691DOH2.PS
DEVICE 2 POSTSCRIPT
CALL E691DOH2.DP
DEVICE 2 CLOSE
SET IPL1NA E691DOK1.PS
DEVICE 2 POSTSCRIPT
CALL E691DOK1.DP
DEVICE 2 CLOSE
SET IPL1NA E691DOK2.PS
DEVICE 2 POSTSCRIPT
CALL E691DOK2.DP
DEVICE 2 CLOSE
SET IPL1NA E691PLO7.PS
DEVICE 2 POSTSCRIPT
CALL E691PLO7.DP
DEVICE 2 CLOSE
END OF CAPTURE
This program will generate the E691 tables and graphs in a
Latex file (E691.TEX).
Date created: 04/19/2005 |
Last updated: 05/07/2024 Please email comments on this WWW page to [email protected]. | ||||||||||||||||||||||||||||||||||||||||||||||